 |
Centre for Australian National Biodiversity
Research
|
Tristan Reekie
The aim of this project was to investigate the evolution of the life cycles of rusts. It was hypothesized that the rusts that undergo an autoecious life cycle evolved from those of the heteroecious life cycles. To investigate the aim a phylogenetic approach was taken.
Devastation of crop by rust  |
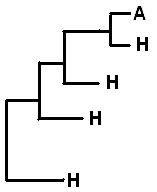
Rusts have complex life cycles. They have up to 5 different spore types during the life cycle produced by different morphological structures. An autoecious life cycle (A) applies to rusts that complete the whole life cycle on one host. For example Melampsora lini, completes its entire life cycle on flax (Linum).
Heteroecious rusts (H) require two often phylogenetically distant hosts, to complete the life cycle. A common example of a heteroecious life cycle for the Melampsora genus is M. epitea, which over winters on Larix while the willow serves as host during summer.
The Melampsora genus was used in this project because both the autoecious and the heteroecious life cycles are found in this genus. Furthermore previous studies have shown that this genus is monophyletic. Molecular tools have been developed for this genus. There are 90 described species in the genus. The species are distributed world wide with only one known to be native to Australia, (M. lini on native flax) while approximately eight others have been introduced to Australia.
Sampling
In total there were 40 samples including 22 species in the collection. Prof. Lars Ericson supplied material collected from Northern Europe John Walker supplied material from Australia, and two samples were collected from around Canberra: M. hypericorum on Hypericum sp.(collected in December 2004) and Melampsora sp. on Salix sp (collected from ANU campus January 2005).
DNA extraction
Two methods were used for DNA extraction:
1. Spores were picked from the leaf material under a microscope using a needle, placed on a microscope slide and crushed between two slides. The crushed spore material was then used for DNA extractions using the Qiagen plant DNeasy purification kit.
2. For the ethanol preserved material from Europe the CTAB method was used.
Polymerase Chain Reaction

The polymerase chain reaction (PCR) was used to amplify the ß-tubulin and translation elongation factors 1α (TEF) genes. A double PCR was carried out using nested primers in the second PCR. The ß-tubulin gene codes for part of the tubulin protein, a dimer formed with α-tubulin. Due to the DNA extraction method used the presence of plant DNA along side rust DNA, required specific primers to ensure that the plant DNA was not amplified. The primers B-tub1317F and B-tub2662R were used for the first touch down PCR while the next PCR was conducted using B-tub 1442F and B-tub2662R or B-tub 3F and B-tub 2662R with the annealing temperature at a constant 55.5° C. A fragment of approximately 1100bp was amplified and purified using the Qiagen PCR purification kit.
Microtubule formed by tubulin  |
For the TEF amplification a double PCR was again used. In the first PCR EF1-526F and Ef1-1567R was used. The nested primers, EFpucc1F and EFbasidR was used. The region amplified with these primers includes three introns. TEF PCR products were gel purified using the Qiagen gel purification kit.
Sequencing and Data Analysis
After cycle sequencing was carried out the product was sent to the John Curtin Medical School for sequencing. Forward and reverse sequences were used to construct contiguous sequences using the program Sequencher. BioEdit and ClustalX was used fro alignment. Two trees, a distance and a parsimony tree, were constructed using PAUP*.
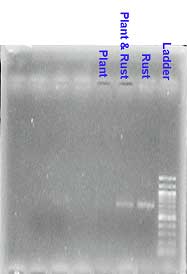 As expected the sequence data showed that most variation between species occurs in the introns. The primers used for ß-tubulin did not amplify plant DNA. Twenty sequences representing 14 species were obtained for the ß-tubulin gene while 28 sequences (with 15 species) for the elongation factor were obtained. Species from a wide range were represented among the sequences obtained. Reasons for non-amplification may include quality of DNA, extraction methods and primers not annealing to the template DNA.
As expected the sequence data showed that most variation between species occurs in the introns. The primers used for ß-tubulin did not amplify plant DNA. Twenty sequences representing 14 species were obtained for the ß-tubulin gene while 28 sequences (with 15 species) for the elongation factor were obtained. Species from a wide range were represented among the sequences obtained. Reasons for non-amplification may include quality of DNA, extraction methods and primers not annealing to the template DNA.
The two trees that are shown (figure 1) are distance trees with bootstrap values showing the support for the nodes. Two main clades are found in both trees (blue and yellow) with the more derived clade (indicated with yellow figure 1) divided into two clades (pink and green)
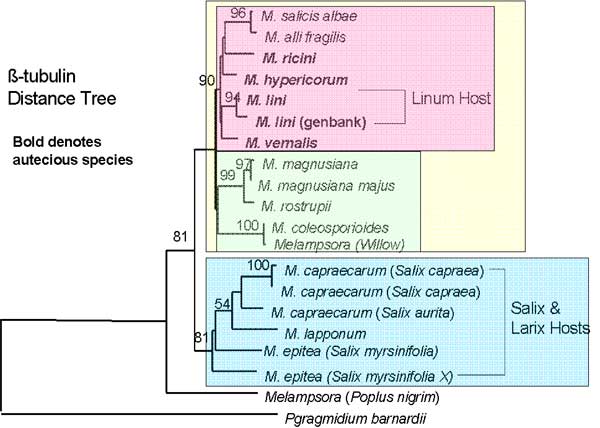
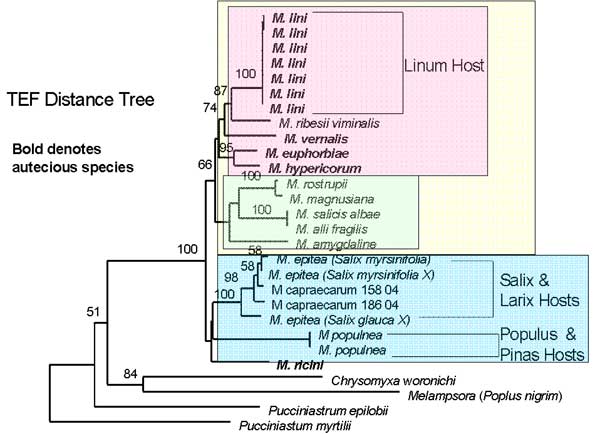
Figure 1. Two distance trees constructed using beta-tubulin and elongation factor gene sequences for Melampsora species.
The phylogenetic trees give more information than just the evolution of the life cycles. By looking at the trees it can be seen that species with the same type of host and life cycles are grouped into the same clade. In one group (blue figure 1) all species are heteroecious with the primary host being an Angiosperm while the alternate host is a Gymnosperm. The second group (yellow group) has both life cycles with all hosts being Angiosperms. Within this only angiosperm host clade the most derived clade (pink) contains all the sampled autoecious species with the heteroecious species as a sister clade. As this clade is most derived it may suggest a possible evolution of the autoecious species from the heteroecious species. Further statistical analysis is necessary to confirm this.
The tree also shows that members of the same species with different hosts are genetically different possibly owing to the fact that they are adapted to these different hosts. Both these points support the theory of co-evolution between plants and rusts. Inoculation studies could be used to determine the specificity of rusts to host. Many M. lini samples were sequenced for TEF because the samples were ecologically different. The molecular work has shown similarities between species previously thought to be unrelated.
In conclusion the results support the hypothesis that autoecious species evolved from heteroecious species.
I would like to thank many people for their help during this project. The Herbarium staff, with special thanks to the Molecular Systematics Lab Staff for all their help and input.
CSIRO Plant Industry, The Centre for Plant Biodiversity and research, and the Pastoral Research Trust for funding and organising the project. And a special thank you to my supervisor Marlien Van Der Merwe for making this all possible.
Ayliffe, M, Dodds, P, Lawrence, G, 2001: Characterisation of a ß-tubulin gene from Melampsora lini and comparison of fungal ß-tubulin genes Mycologia Res. 105(7)
McAlpine, D, 1906: The rusts of Australia: their structure, nature and classification, Melbourne, Govt. Printer
Petersen, R, 1974: The Rust Fungus Life Cycle, The Botanical Review 40(4) 453-513
Smith, J, Blanchette, R, Newcombe, G, 2004: Molecular and morphological characterization of the willow rust fungus, Melampsora epitea, from artic and temperate hosts in North America, Mycologia 96(6) 1330-1338
Tian, C, Shang, Y, Zhuang J, Wang, Q, Kakishima, M, 2004: Morphological and molecular phylogenetic analysis of Melampsora species on poplars in China, Mycoscience 45 56-66